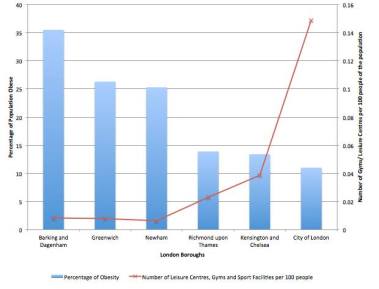
During our research, we were particularly interested in investigating the impact of the environment on a population’s lifestyle. We were eager to various assumptions and correlations, and hence simulated choropleth maps to visualise the relationship between obesity and various lifestyle-affecting factors.
Although there are both advantages and disadvantages when using a map for data visualisation, in this case, not reflecting the population size, they are nevertheless a valuable tool when demonstrating various geographical patterns, and in comparison, when investigating connections between different factors.
Initially we made maps using python. Although these maps provided some insight into the subject matter, Python had too many limitations to use for the final visualisation. Therefore, we decided to use Carto, which solved many of the occurring problems in Python. In this section, web have recorded the process and method for both Python and Carto.
Python
After cleaning the data, we were able to use the code given in the QM workshop 8 to simulate various choropleth maps. Through interacting with the code, and further modifying it, these maps became a useful way of illustrating various data per factor. As the geography data used in the workshop was London by ward, we had to find the data for London boroughs. The first challenge was to use a format supported by geopandas. We had to ensure that all the files with different extensions were in order, to ascertain the functionality of all the shp file.
To merge the files, we used the code learned in class, which allows merging using a column that is shared by both files. To do this properly, the name of the column had to be renamed so that the equivalent values were under the same column name.

After merging the data of each factor (such as fast food) with the geography data, which was relatively confusing due to the various column names and sections, it was relatively straightforward. By visualising and saving the image file, we were able to produce a map for each of the factors.

The difficulty of Python is the configuration of the map to fit our needs. Although changing the color scheme is relatively easy, deciding the threshold for a particular color was too difficult for us, especially as the code for using quantiles was devised by someone online, which meant it was necessary to programme according to this code. The reason why it is necessary to change thresholds is to make sure that comparison between maps can be done in the most effective way. It does not make sense to compare a map that is mostly one color and one that is very varied. Quantiles can help solve this problem, but the question remains whether useful comparisons can be made between different factors with different mean values. For example, how can we ensure that a choropleth of fast food outlets, whose average is generally over a thousand, and a choropleth of gym facilities, which are generally less than a thousand, give a good comparison? Quantiles are dividing the color levels according to the values from the data. However, it might be more fitting to set our own levels at which we think is most beneficial to color them.
Carto
When using Carto, the problem we encountered involved correct formatting of the file; uploading CSV files did not work, and we had to upload instead a folder with the ship file in it, deleting the unnecessary files, but keeping the same files in different formats (different extensions). After uploading the file, it was also necessary to manually imput data by adding new columns. The dashboard tools allows easy manipulation of colors.
The advantage with Carto is that the legend, title, and intervals can be manipulated by us. In addition, there is a better user interface as anyone who uses the map may zoom in and drag the map to view what is most interesting, as well as see the actual values for each borough by moving the cursor onto the map. The color scheme for all maps are the same, to enable easier and quicker comparison between them.